|
3/1/2024 0 Comments Paper out in Science
0 Comments
1/31/2024 0 Comments Slow to respond for a bit...Happy to announce that today I completed a 9 month, in-depth study of vertebrate embryology and development. Thankfully N=1, but I will likely not be getting sleep for the next year.
After a fantastic SICB symposium on marine larval genomics, I'm happy to share a publication focused on how to improve our genome publications. Working on marine systems, we know that non-model organisms present challenges that require innovation to overcome. The more detail we include in our work, the better we aid our community of scientists to move forward. The checklist is designed to ensure that each stage of a genome project is documented thoroughly, from the animal collection to the final data availability. Formatted as a fillable PDF, the goal is to help researchers produce publications with the most complete methods sections possible, and to help reviewers to be thorough when reading manuscripts that may be on organisms they are unfamiliar with. Here's hoping that this becomes a useful resource!
Check it out HERE!
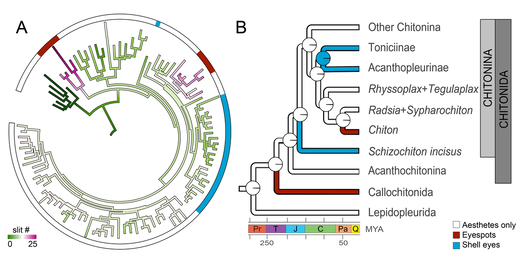
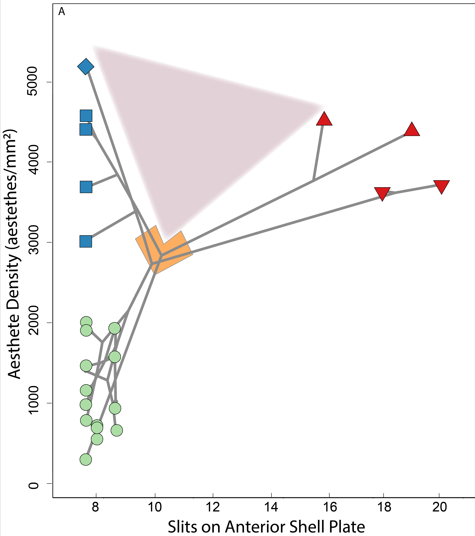
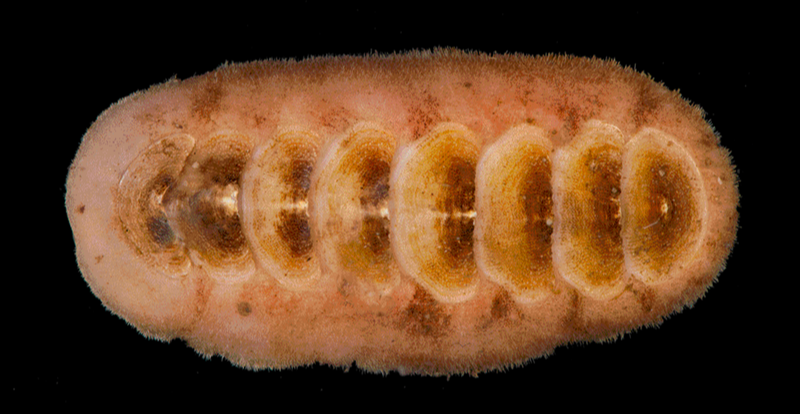
We investigated the morphology of both types of eyes, and found a difference in the shells on the chitons' backs. Shell eyes and eyespots are embedded in the upper layer of chiton shells, so the optic nerves have to travel to an edge where they pass through openings (slits) to get down to the rest of the chiton's body and join the big nerve cords. We found that chitons with eyespots have way more openings than chitons with shell eyes, or with no eyes. Why? Well, you need hundreds of shell eyes but THOUSANDS of eyespots, so we think that more openings work kind of like a cable organizer at the back of your desk, helping keep all those optic nerves straight. Because eyespots work together, chitons can't let them get all tangled up without losing 'sight' of which eye is where!
Had a delightful (but EARLY) morning presenting on some new chiton work as part of the Tangled Bank seminar series in Evolution! It was so fun to share this work -- stay tuned for a preprint very soon! -- and I had the joy of receiving a lot of fantastic questions. That said, I probably WON'T give another seminar at 5am my time. At least I didn't yawn in the middle of my own talk!
8/30/2022 0 Comments New Sea Spider Phylogenetics Paper
7/25/2022 0 Comments Back from PanamaSummer sampling travels continued! Todd, Cheyenne, Niko and I just got back from Panama, where we spent 2.5 weeks working at STRI in Bocas. Emily and Bridget took a course in Cnidarians/Ctenophores, so we had a fantastic time as a lab hitting the water and watching ostracods signal. The species resident in the sea grass beds there signals synchronously, so we got to watch waves and waves of spectacular bioluminescence. I can't wait to dive into the transcriptomes from the many ostracods we get to bring home. We also tried a mark-recapture of ostracods; we actually DID recapture some, which surprised all of us! And of course, there were sloths.
6/28/2022 0 Comments Back from Puerto Rico
5/23/2022 0 Comments New chiton genome paper!
|









 RSS Feed
RSS Feed